Welcome
Computational Tools for Macromolecular Neutron Crystallography (MNC) is a consortium between Oak Ridge National Laboratory(ORNL) and Lawrence Berkeley National Laboratory (LBNL). Our aim has been to address the need for new computational tools and methods to deal with the increasing number of neutron macromolecular structures being determined and their increasing size and complexity.
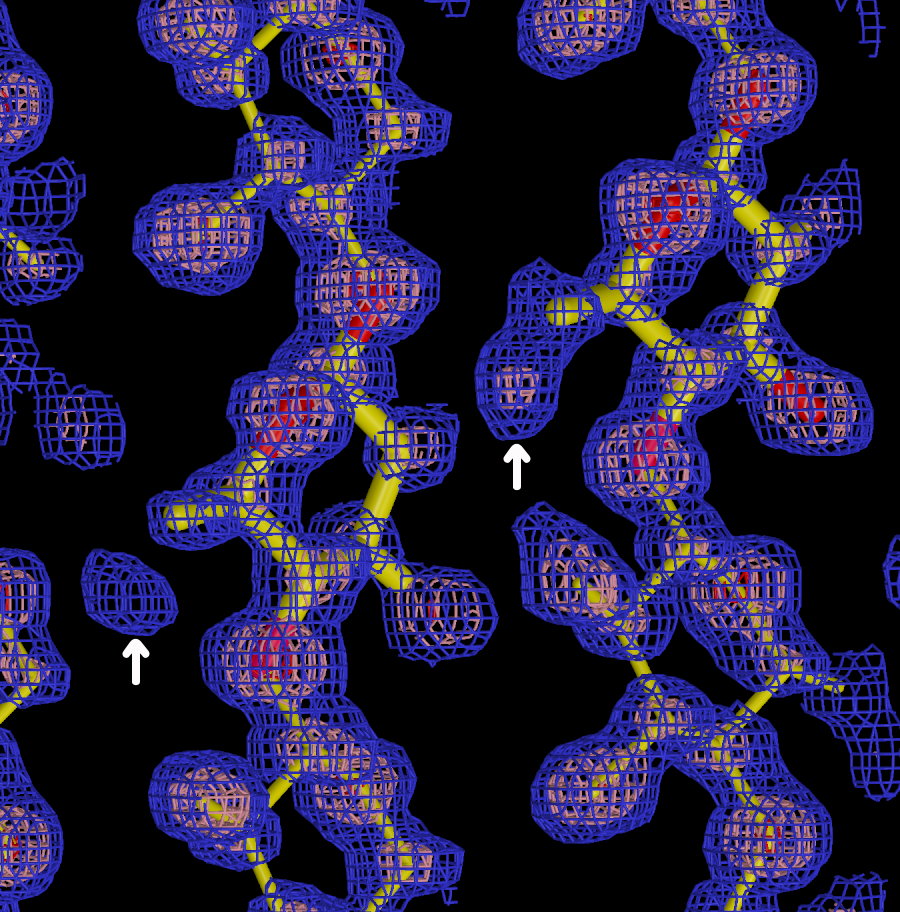
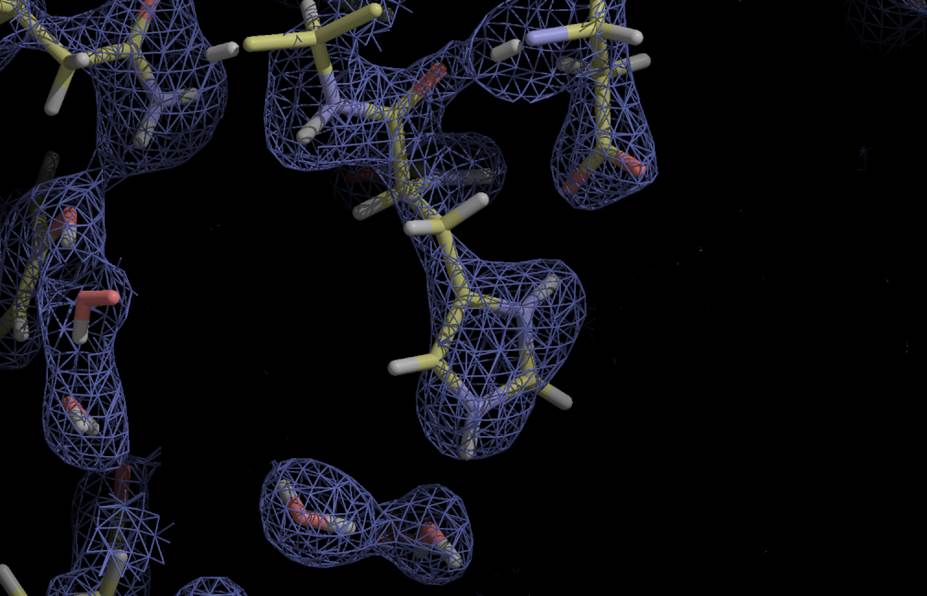
Since 2004 we have developed improved algorithms for the refinement of models against neutron diffraction data with the nCNS program (Adams et al., 2009) and phenix.refine within the Phenix package (Afonine et al., 2010). Over 60% of deposited neutron structures have made use of our software. Since 2007, 78% of neutron structures deposited have used our software, in particular phenix.refine. In the last 4 years we have focused our efforts on implementing new algorithms for neutron structure determination in the Phenix package, as this is very actively developed and widely used in the crystallographic community (over 3000 PDB depositions in 2013, and over 1200 citations of the Phenix software in 2013). The development and distribution of nCNS and Phenix for global structure refinement using generalized neutron, X-ray and energy refinement, which we call XN refinement, has removed a critical barrier to progress in this field, as there were several difficulties associated with refining neutron structures using previous software and approaches. However, in our interactions with the growing neutron crystallography user community it is becoming increasingly clear that there are remaining challenges that limit progress in this field and that will require the innovative solutions proposed in this project.
The macromolecular structures determined using neutrons so far have been medium-sized proteins and nucleic acids that can be prepared as relatively large crystals, similar to those studied by X-rays in the first decade of the PDB. However, experimental facilities are expanding the capabilities of neutron crystallography to new kinds of biological questions. The computational tools developed in this project will be used to shift current research to more complex structures that are larger in size, complexity and biomedical relevance. This shift in the scope of neutron research will require developing new computational concepts and approaches. We will develop a novel approach of data processing that makes use of knowledge about the atomic model to improve the accuracy of marginal weak data obtained from more complex systems. Our innovative approach is inspired by Rietveld refinement, and may also benefit X-ray studies of complex systems with large unit cells or high mosaicity and disorder. We will adapt and apply several approaches that have been developed to improve electron density maps calculated from weak or incomplete X-ray data to neutron density maps for the first time. Neutron density maps have both significant positive (O, C, N, P) and negative (H) features, so this will involve developing new approaches and solutions. We will design and implement a new approach to modeling bulk solvent that takes into account its non-uniform nature. This will be important for neutron studies of membrane proteins with bulk solvent that includes lipid, detergent, and crystallizing agents which may introduce heterogeneity. Finally we will explore the first application of ensemble refinement techniques to neutron crystallography to better model dynamics and disorder and extract more meaningful solvation models. For neutron studies we need to implement improved force fields that realistically model the interactions of protein and solvent in the time averaged ensemble structure refinement.
These innovations will open up a larger set of biological problems that can be addressed with neutrons. Complexes containing proteins and other biological molecules will be studied to determine the role of solvent and H-bonding in mediating interactions. A number of extremophile Archaea and bacterial species utilize unusual metabolic pathways and rely on novel chemistry to sustain life, and neutrons can reveal this new chemistry. Membrane proteins are a rich target for neutron crystallography, and neutrons can potentially offer insight into the chemical environment within pores and channels, and the molecular basis of ion selectivity. A detailed understanding of structure and mechanism through neutron crystallography can guide future drug development efforts targeting membrane proteins.